Implementing K-Means
ML Jähnichen HUB
prerequisites
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
Data
def old_faithful():
filename = "data.tsv"
# read file
df = pd.read_csv(filename, delimiter=r"\s+", skiprows=0, header=0)
# delete unimportant columns
df = df.drop(['idx'], axis=1)
# normalize
normal = df.as_matrix()
normal = (normal - normal.min(axis=0))/normal.ptp(axis=0)
return df.as_matrix(), normal
data, _data = old_faithful()
Visualization
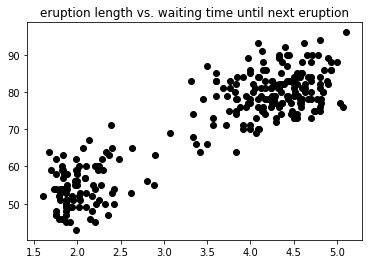
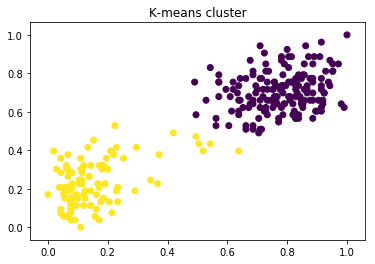
def vis(data, assign='black'):
if assign is 'black':
title = "eruption length vs. waiting time until next eruption"
else:
title = "K-means cluster"
plt.scatter(data[:, 0], data[:, 1], c=assign)
plt.title(title)
plt.show()
vis(data)
Clustering
class KMeans:
"""
Implement K-means algorithm
:param K: hyperparameter - number of clusters
:param X: data numpy matrix, every row one data point
"""
def __init__(self, K, X):
self.K = K
self.X = X
def __len__(self):
"""number of data points"""
return len(self.X)
def fit(self):
# place cluster centers randomly for all K
centres = np.random.uniform(0, +1, size=(self.K, 2)) # cluster centres
infer = np.zeros(len(self)) # vector with corresponding cluster for X
while True:
_centres = centres.copy()
for i, p in enumerate(self.X): # assign datapoints to nearest cluster
# distance from points and cluster centers
_d = np.linalg.norm(centres - p, axis=1)
# get indice with smallest distance
infer[i] = _d.argmin()
# todo filter nan
for j in range(self.K): # recompute cluster centres
_center = np.mean(self.X[infer == j])
if _center == _center: # nan check
centres[j] = _center
if np.max(centres-_centres) < 1e-8: # check convergence
break
return infer, centres
kmean = KMeans(2, _data)
label, _ = kmean.fit()
vis(_data, label)
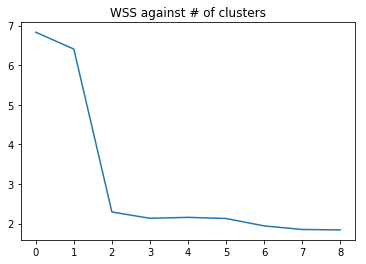
within-cluster sum of squares
def wss(data, centres, labels):
norm = np.linalg.norm(centres[labels.astype('int')] - data)
sum = np.sum(norm)
return sum
def b(bound=10):
measures = np.zeros(bound -1)
for i in range(1, bound):
kmean = KMeans(i, _data)
labels, centres = kmean.fit()
measures[i-1] = wss(_data, centres, labels)
plt.plot(measures)
plt.title("WSS against # of clusters")
plt.show()
b()
/home/float/.local/lib/python3.5/site-packages/numpy/core/fromnumeric.py:2909: RuntimeWarning: Mean of empty slice.
out=out, **kwargs)
/home/float/.local/lib/python3.5/site-packages/numpy/core/_methods.py:80: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
 Music
Music